上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
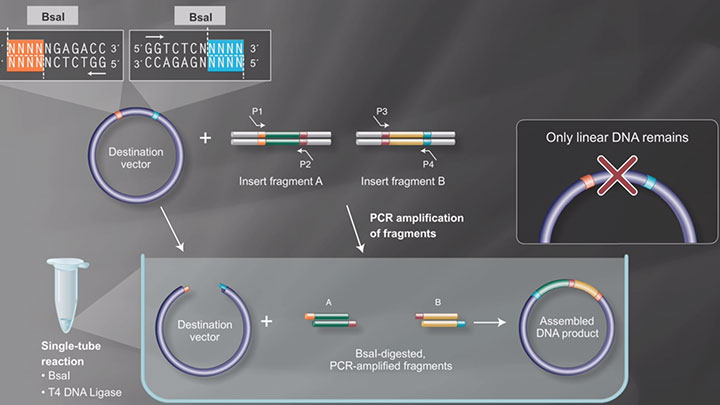
BsaI
This product was discontinued on 12/31/2020 and replaced with the High Fidelity version, BsaI-HFv2 (NEB #R3733).
- Catalog # R0535 was discontinued on December 31, 2020
Attention Golden Gate Assembly users: BsaI-HFv2 (NEB #R3733) has been optimized for Golden Gate Assembly*. BsaI-HFv2 also works well for any protocol requiring DNA cutting by BsaI. This is the recommended enzyme for any purpose requiring digestion at the recognition sequence …5′-GGTCTC(N1)/(N5)-3′…
*The requirements for digestion of DNA during Golden Gate Assembly are more demanding than what is encountered in traditional DNA cloning. For Golden Gate, the enzyme must function well in a buffer that might not be the same buffer as that normally provided for the restriction enzyme, and maintain activity for an extended time at elevated temperatures in a dynamic cutting/re-ligating reaction with competition for substrate binding between the endonuclease and ligase. Extensive testing has demonstrated superior performance of BsaI-HFv2 compared to both BsaI (NEB #R0535) and BsaI-HF (NEB
- High Fidelity (HF®) version available () in this challenging Golden Gate assembly context where restriction enzyme efficiency and fidelity are critically important.“/products/r3733-bsai-hf-v2″>NEB #R3733) supplied with CutSmart® Buffer
- Used in Golden Gate Assembly
- Type IIS restriction enzymes recognize asymmetric DNA sequences and cleave outside of their recognition sequence
- 100% activity in CutSmart Buffer (over 210 enzymes are available in the same buffer) simplifying double digests
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
精选视频
产品信息
High Fidelity (HF) Restriction Enzymes have 100% activity in CutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
产品来源
An E. coli strain that carries the cloned BsaI gene from Bacillus stearothermophilus 6-55 (Z. Chen)
- 产品类别:
- Discontinued Products
- 特性和用法
单位定义
One unit is defined as the amount of enzyme required to digest 1 µg of pXba DNA in 1 hour at 37°C in a total reaction volume of 50 µl.
反应条件
1X CutSmart® Buffer
Incubate at 37°C1X CutSmart® Buffer
50 mM Potassium Acetate
20 mM Tris-acetate
10 mM Magnesium Acetate
100 µg/ml BSA
(pH 7.9 @ 25°C)在不同缓冲液中的活性
NEBuffer™ 1.1: 75%
NEBuffer™ 2.1: 75%
NEBuffer™ 3.1: 100%稀释兼容性
- 稀释液 B
贮存溶液
10 mM Tris-HCl
300 mM NaCl
1 mM DTT
0.1 mM EDTA
500 µg/ml BSA
50% Glycerol
pH 7.4 @ 25°C热失活
65°C for 20 min
甲基化敏感性
dam 甲基化: Not Sensitive
dcm 甲基化: Impaired by Some Combinations of Overlapping
CpG甲基化: Blocked by Some Combinations of Overlapping - 相关产品
相关产品
- dam–/dcm– Competent E. coli
- Monarch® Plasmid Miniprep Kit
- Monarch® DNA Gel Extraction Kit
- Monarch® PCR & DNA Cleanup Kit (5 μg)
单独销售的组分
- Gel Loading Dye, Purple (6X)
- CutSmart® Buffer
- 注意事项
- Blocked by overlapping dcm methylation. Cleavage of mammalian genomic DNA is blocked by some combinations of overlapping CpG methylation.
- Activity at 50°C is 100%.
- May exhibit star activity in NEBuffer r1.1.
- Based on the stability of the enzyme in the reaction, incubations longerthan 1 hr will not result in improved digestion, unless additionalenzyme is added. Please refer to Restriction endonuclease survival in a reaction for more information regarding this topic.
- star activity may result from a glycerol concentration of >5%
操作说明、说明书 & 用法
- 操作说明
- Optimizing Restriction Endonuclease Reactions
- Restriction Digest Protocol
- Double Digest Protocol with Standard Restriction Enzymes
- 使用指南
- Activity at 37°C for Restriction Enzymes with Alternate Incubation Temperatures
- Activity of Restriction Enzymes in PCR Buffers
- Cleavage Close to the End of DNA Fragments
- Dam and Dcm Methylases of E. coli
- Digestion of Agarose-Embedded DNA: Info for Specific Enzymes
- Double Digests
- Effects of CpG Methylation on Restriction Enzyme Cleavage
- Heat Inactivation
- NEBuffer Activity/Performance Chart with Restriction Enzymes
- Optimizing Restriction Endonuclease Reactions
- Restriction Endonucleases – Survival in a Reaction
- Restriction Enzyme Diluent Buffer Compatibility
- Restriction Enzyme Tips
- Single Letter Codes
- Star Activity
- Technical Tips For Optimizing Golden Gate Assembly Reactions
- Traditional Cloning Quick Guide
- 应用实例
- Breaking through the Limitations of Golden Gate Assembly
工具 & 资源
- 选择指南
- Alphabetized List of Recognition Sequences
- Cleavage of Supercoiled DNA
- Compatible Cohesive Ends and Generation of New Restriction Sites
- Dam-Dcm and CpG Methylation
- Enzymes with Nonpalindromic Sequences
- Frequencies of Restriction Sites
- Isoelectric Points (pI) for Restriction Enzymes
- Isoschizomers
- NEB Diluent and Buffer Table
- Time-Saver™ Qualified Enzymes
- Type IIS Restriction Enzymes
- Why Choose Recombinant Enzymes?
- Web 工具
- Competitor Cross-Reference Tool
- DNA Sequences and Maps Tool
- Double Digest Finder
- Enzyme Finder
- NEBcutter™ v3.0
- NEBioCalculator®
- REBASE®
FAQs & 问题解决指南
- FAQs
- Which restriction enzymes are used in Golden Gate Assembly?
- Which restriction enzymes are used in GoldenBraid Assembly?
- Why is my Restriction Enzyme not cutting DNA?
- Why do I see a DNA smear on an agarose gel after a restriction digest?
- Why do I see additional DNA bands on my gel after a restriction digest?
- How many nucleotides do I have to add adjacent to the RE recognition site in order to get efficient cutting?
- Which NEB restriction enzymes are supplied with Gel Loading Dye, Purple (6X)?
- What is the activity of the Type IIS restriction enzyme BsaI-HFv2 (NEB #R3733) in T4 DNA Ligase Buffer?
- Is this enzyme sensitive to dam, dcm or mammalian CpG methylation?
- Can Gel Loading Dye, Purple 6X (B7024) be stored in cold temperatures?
- 问题解决指南
- Restriction Enzyme Troubleshooting Guide
- 实验技巧
- Restriction Enzymes in Golden Gate Assembly

- Restriction Enzymes in Golden Gate Assembly
引用 & 技术文献
- 产品手册
- Golden Gate Assembly brochure
- 引用文献
产品引用文献查找工具
 Powered by Bioz See more details on Bioz
Powered by Bioz See more details on Bioz更多引用文献
- Lee JH, Won HJ, Oh E-S, Oh M-H and Jung JH (2020) Golden Gate Cloning-Compatible DNA Replicon/2A-Mediated Polycistronic Vectors for Plants Front Plant Sci; 11, 559365. DOI: 10.3389/fpls.2020.559365
- Feng Y, Zhang S, Huang X (2014) A robust TALENs system for highly efficient mammalian genome editing Sci Rep; 4, 3632. PubMedID: 24407151, DOI: 10.1038/srep03632
- Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system Sci Rep; 4, 5400. PubMedID: 24954249, DOI: 10.1038/srep05400
质控、安全 & 法规
- 质控分析
每一新批次的 NEB 产品都要经过质控,以满足指定的质量标准,对特定产品的质量标准和每一批次的检测数据可以通过产品说明表格、COA、产品信息卡或者产品手册进行查阅或下载。关于 NEB 产品质控的详细信息,可从此处查阅。
- 产品说明与变更通知
产品说明与变更通知
产品说明表包含产品的储存温度、有效期和规格,这些文件的命名规则如下:[货号]_[规格]_[版本]
- R0535S_L_v2
- CoA
CoA 文件包含单个批次产品的储存温度、有效期和质控,这些文件的命名规则如下: [货号]_[规格]_[版本]_[Lot#]
- R0535S_L_v1_0371403
- R0535S_L_v1_0361212
- R0535S_L_v1_0361305
- R0535S_L_v1_0361311
- R0535S_L_v1_0371404
- R0535S_L_v2_0371404
- R0535S_L_v1_0371409
- R0535S_L_v2_0371503
- R0535S_L_v2_0371509
- R0535S_L_v2_0371603
- R0535S_L_v2_0381608
- R0535S_L_v2_0401702
- R0535S_L_v2_0401708
- R0535S_L_v2_0431712
- R0535S_L_v2_0431805
- R0535L_v2_10009918
- R0535S_v2_10018570
- R0535L_v2_10030542
- R0535S_v2_10030544
- R0535S_v2_10043185
- R0535L_v2_10043183
- R0535S_v2_10055060
- R0535S_v2_10059599
- R0535S_v2_10062820
- R0535L_v2_10062822
- R0535S_v2_10065817
- R0535S_v2_10081423
- R0535S_v2_10091140
- R0535L_v2_10086217
- SDS
以下 SDS 文件可以帮助您安全地使用该产品
-
BsaI
-
BsaI |NEB酶试剂 New England Biolabs
上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
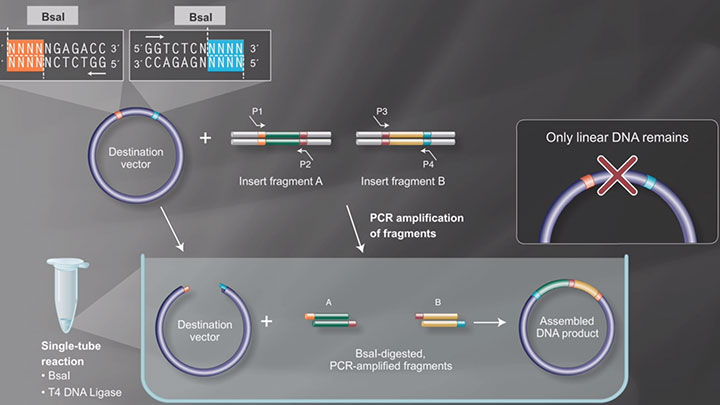
BsaI
This product was discontinued on 12/31/2020 and replaced with the High Fidelity version, BsaI-HFv2 (NEB #R3733).
- Catalog # R0535 was discontinued on December 31, 2020
Attention Golden Gate Assembly users: BsaI-HFv2 (NEB #R3733) has been optimized for Golden Gate Assembly*. BsaI-HFv2 also works well for any protocol requiring DNA cutting by BsaI. This is the recommended enzyme for any purpose requiring digestion at the recognition sequence …5′-GGTCTC(N1)/(N5)-3′…
*The requirements for digestion of DNA during Golden Gate Assembly are more demanding than what is encountered in traditional DNA cloning. For Golden Gate, the enzyme must function well in a buffer that might not be the same buffer as that normally provided for the restriction enzyme, and maintain activity for an extended time at elevated temperatures in a dynamic cutting/re-ligating reaction with competition for substrate binding between the endonuclease and ligase. Extensive testing has demonstrated superior performance of BsaI-HFv2 compared to both BsaI (NEB #R0535) and BsaI-HF (NEB
- High Fidelity (HF®) version available () in this challenging Golden Gate assembly context where restriction enzyme efficiency and fidelity are critically important.“/products/r3733-bsai-hf-v2″>NEB #R3733) supplied with CutSmart® Buffer
- Used in Golden Gate Assembly
- Type IIS restriction enzymes recognize asymmetric DNA sequences and cleave outside of their recognition sequence
- 100% activity in CutSmart Buffer (over 210 enzymes are available in the same buffer) simplifying double digests
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
精选视频
产品信息
High Fidelity (HF) Restriction Enzymes have 100% activity in CutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
产品来源
An E. coli strain that carries the cloned BsaI gene from Bacillus stearothermophilus 6-55 (Z. Chen)
- 产品类别:
- Discontinued Products
- 特性和用法
单位定义
One unit is defined as the amount of enzyme required to digest 1 µg of pXba DNA in 1 hour at 37°C in a total reaction volume of 50 µl.
反应条件
1X CutSmart® Buffer
Incubate at 37°C1X CutSmart® Buffer
50 mM Potassium Acetate
20 mM Tris-acetate
10 mM Magnesium Acetate
100 µg/ml BSA
(pH 7.9 @ 25°C)在不同缓冲液中的活性
NEBuffer™ 1.1: 75%
NEBuffer™ 2.1: 75%
NEBuffer™ 3.1: 100%稀释兼容性
- 稀释液 B
贮存溶液
10 mM Tris-HCl
300 mM NaCl
1 mM DTT
0.1 mM EDTA
500 µg/ml BSA
50% Glycerol
pH 7.4 @ 25°C热失活
65°C for 20 min
甲基化敏感性
dam 甲基化: Not Sensitive
dcm 甲基化: Impaired by Some Combinations of Overlapping
CpG甲基化: Blocked by Some Combinations of Overlapping - 相关产品
相关产品
- dam–/dcm– Competent E. coli
- Monarch® Plasmid Miniprep Kit
- Monarch® DNA Gel Extraction Kit
- Monarch® PCR & DNA Cleanup Kit (5 μg)
单独销售的组分
- Gel Loading Dye, Purple (6X)
- CutSmart® Buffer
- 注意事项
- Blocked by overlapping dcm methylation. Cleavage of mammalian genomic DNA is blocked by some combinations of overlapping CpG methylation.
- Activity at 50°C is 100%.
- May exhibit star activity in NEBuffer r1.1.
- Based on the stability of the enzyme in the reaction, incubations longerthan 1 hr will not result in improved digestion, unless additionalenzyme is added. Please refer to Restriction endonuclease survival in a reaction for more information regarding this topic.
- star activity may result from a glycerol concentration of >5%
操作说明、说明书 & 用法
- 操作说明
- Optimizing Restriction Endonuclease Reactions
- Restriction Digest Protocol
- Double Digest Protocol with Standard Restriction Enzymes
- 使用指南
- Activity at 37°C for Restriction Enzymes with Alternate Incubation Temperatures
- Activity of Restriction Enzymes in PCR Buffers
- Cleavage Close to the End of DNA Fragments
- Dam and Dcm Methylases of E. coli
- Digestion of Agarose-Embedded DNA: Info for Specific Enzymes
- Double Digests
- Effects of CpG Methylation on Restriction Enzyme Cleavage
- Heat Inactivation
- NEBuffer Activity/Performance Chart with Restriction Enzymes
- Optimizing Restriction Endonuclease Reactions
- Restriction Endonucleases – Survival in a Reaction
- Restriction Enzyme Diluent Buffer Compatibility
- Restriction Enzyme Tips
- Single Letter Codes
- Star Activity
- Technical Tips For Optimizing Golden Gate Assembly Reactions
- Traditional Cloning Quick Guide
- 应用实例
- Breaking through the Limitations of Golden Gate Assembly
工具 & 资源
- 选择指南
- Alphabetized List of Recognition Sequences
- Cleavage of Supercoiled DNA
- Compatible Cohesive Ends and Generation of New Restriction Sites
- Dam-Dcm and CpG Methylation
- Enzymes with Nonpalindromic Sequences
- Frequencies of Restriction Sites
- Isoelectric Points (pI) for Restriction Enzymes
- Isoschizomers
- NEB Diluent and Buffer Table
- Time-Saver™ Qualified Enzymes
- Type IIS Restriction Enzymes
- Why Choose Recombinant Enzymes?
- Web 工具
- Competitor Cross-Reference Tool
- DNA Sequences and Maps Tool
- Double Digest Finder
- Enzyme Finder
- NEBcutter™ v3.0
- NEBioCalculator®
- REBASE®
FAQs & 问题解决指南
- FAQs
- Which restriction enzymes are used in Golden Gate Assembly?
- Which restriction enzymes are used in GoldenBraid Assembly?
- Why is my Restriction Enzyme not cutting DNA?
- Why do I see a DNA smear on an agarose gel after a restriction digest?
- Why do I see additional DNA bands on my gel after a restriction digest?
- How many nucleotides do I have to add adjacent to the RE recognition site in order to get efficient cutting?
- Which NEB restriction enzymes are supplied with Gel Loading Dye, Purple (6X)?
- What is the activity of the Type IIS restriction enzyme BsaI-HFv2 (NEB #R3733) in T4 DNA Ligase Buffer?
- Is this enzyme sensitive to dam, dcm or mammalian CpG methylation?
- Can Gel Loading Dye, Purple 6X (B7024) be stored in cold temperatures?
- 问题解决指南
- Restriction Enzyme Troubleshooting Guide
- 实验技巧
- Restriction Enzymes in Golden Gate Assembly

- Restriction Enzymes in Golden Gate Assembly
引用 & 技术文献
- 产品手册
- Golden Gate Assembly brochure
- 引用文献
产品引用文献查找工具
 Powered by Bioz See more details on Bioz
Powered by Bioz See more details on Bioz更多引用文献
- Lee JH, Won HJ, Oh E-S, Oh M-H and Jung JH (2020) Golden Gate Cloning-Compatible DNA Replicon/2A-Mediated Polycistronic Vectors for Plants Front Plant Sci; 11, 559365. DOI: 10.3389/fpls.2020.559365
- Feng Y, Zhang S, Huang X (2014) A robust TALENs system for highly efficient mammalian genome editing Sci Rep; 4, 3632. PubMedID: 24407151, DOI: 10.1038/srep03632
- Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system Sci Rep; 4, 5400. PubMedID: 24954249, DOI: 10.1038/srep05400
质控、安全 & 法规
- 质控分析
每一新批次的 NEB 产品都要经过质控,以满足指定的质量标准,对特定产品的质量标准和每一批次的检测数据可以通过产品说明表格、COA、产品信息卡或者产品手册进行查阅或下载。关于 NEB 产品质控的详细信息,可从此处查阅。
- 产品说明与变更通知
产品说明与变更通知
产品说明表包含产品的储存温度、有效期和规格,这些文件的命名规则如下:[货号]_[规格]_[版本]
- R0535S_L_v2
- CoA
CoA 文件包含单个批次产品的储存温度、有效期和质控,这些文件的命名规则如下: [货号]_[规格]_[版本]_[Lot#]
- R0535S_L_v1_0371403
- R0535S_L_v1_0361212
- R0535S_L_v1_0361305
- R0535S_L_v1_0361311
- R0535S_L_v1_0371404
- R0535S_L_v2_0371404
- R0535S_L_v1_0371409
- R0535S_L_v2_0371503
- R0535S_L_v2_0371509
- R0535S_L_v2_0371603
- R0535S_L_v2_0381608
- R0535S_L_v2_0401702
- R0535S_L_v2_0401708
- R0535S_L_v2_0431712
- R0535S_L_v2_0431805
- R0535L_v2_10009918
- R0535S_v2_10018570
- R0535L_v2_10030542
- R0535S_v2_10030544
- R0535S_v2_10043185
- R0535L_v2_10043183
- R0535S_v2_10055060
- R0535S_v2_10059599
- R0535S_v2_10062820
- R0535L_v2_10062822
- R0535S_v2_10065817
- R0535S_v2_10081423
- R0535S_v2_10091140
- R0535L_v2_10086217
- SDS
以下 SDS 文件可以帮助您安全地使用该产品
-
BsaI
-
EagI | NEB酶试剂 New England Biolabs
上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
EagI
This product has been discontinued and is replaced with the High-Fidelity version, EagI-HF (NEB #R3505)
- Catalog # R0505 was discontinued on March 17, 2021
- Time-Saver™ qualified for digestion in 5-15 minutes
- High Fidelity (HF®) version available (NEB #3505) supplied with CutSmart® Buffer
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
- Restriction Enzyme Cut Site: C/GGCCG
精选视频
产品信息
EagI has a High Fidelity version, EagI-HF® (NEB #R3505)
High Fidelity (HF) Restriction Enzymes have 100% activity in CutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
产品来源
An E. coli strain that carries the EagI gene from Enterobacter agglomerans (R. Morgan).
- 产品类别:
- Discontinued Products
- 特性和用法
单位定义
One unit is defined as the amount of enzyme required to digest 1 µg of pXba DNA in 1 hour at 37°C in a total reaction volume of 50 µl.
反应条件
1X NEBuffer™ 3.1
Incubate at 37°C1X NEBuffer™ 3.1
100 mM NaCl
50 mM Tris-HCl
10 mM MgCl2
100 µg/ml BSA
(pH 7.9 @ 25°C)在不同缓冲液中的活性
NEBuffer™ 1.1: 10%
NEBuffer™ 2.1: 25%
NEBuffer™ 3.1: 100%稀释兼容性
- 稀释液 B
贮存溶液
10 mM Tris-HCl
500 mM NaCl
1 mM DTT
0.1 mM EDTA
200 µg/ml BSA
50% Glycerol
pH 8 @ 25°C热失活
65°C for 20 min
甲基化敏感性
dam 甲基化: Not Sensitive
dcm 甲基化: Not Sensitive
CpG甲基化: Blocked同裂酶
XmaIII+
- 相关产品
相关产品
- EagI-HF®
- Monarch® Plasmid Miniprep Kit
- Monarch® DNA Gel Extraction Kit
- Monarch® PCR & DNA Cleanup Kit (5 μg)
单独销售的组分
- Gel Loading Dye, Purple (6X)
- NEBuffer 3.1
- 注意事项
- EagI is an isoschizomer of XmaIII.
- For full EagI activity, the pH of the reaction mix must be between (7.9 and 9.0 @ 25°C).
- This enzyme has shown to have lower activity on some supercoiled plasmids, with more than 1 unit required to digest 1 μg plasmid DNA. For complete digestion of 1 μg of plasmid DNA please follow our recommended digestion protocol.
- Blocked by CpG methylation.
操作说明、说明书 & 用法
- 操作说明
- Optimizing Restriction Endonuclease Reactions
- Restriction Digest Protocol
- Double Digest Protocol with Standard Restriction Enzymes
- 使用指南
- Activity at 37°C for Restriction Enzymes with Alternate Incubation Temperatures
- Activity of Restriction Enzymes in PCR Buffers
- Cleavage Close to the End of DNA Fragments
- Dam and Dcm Methylases of E. coli
- Digestion of Agarose-Embedded DNA: Info for Specific Enzymes
- Double Digests
- Effects of CpG Methylation on Restriction Enzyme Cleavage
- Heat Inactivation
- NEBuffer Activity/Performance Chart with Restriction Enzymes
- Optimizing Restriction Endonuclease Reactions
- Restriction Endonucleases – Survival in a Reaction
- Restriction Enzyme Diluent Buffer Compatibility
- Restriction Enzyme Tips
- Single Letter Codes
- Star Activity
- Traditional Cloning Quick Guide
工具 & 资源
- 选择指南
- Alphabetized List of Recognition Sequences
- Cleavage of Supercoiled DNA
- Compatible Cohesive Ends and Generation of New Restriction Sites
- Dam-Dcm and CpG Methylation
- Frequencies of Restriction Sites
- Isoelectric Points (pI) for Restriction Enzymes
- Isoschizomers
- NEB Diluent and Buffer Table
- Recleavable Filled-in 5′ Overhangs
- Time-Saver™ Qualified Enzymes
- Why Choose Recombinant Enzymes?
- Web 工具
- Competitor Cross-Reference Tool
- DNA Sequences and Maps Tool
- Double Digest Finder
- Enzyme Finder
- NEBcutter™ v3.0
- NEBioCalculator®
- REBASE®
FAQs & 问题解决指南
- FAQs
- Does EagI produce commonly used compatible ends?
- Are more units of EagI required to cut supercoiled DNA than lambda DNA?
- Do I have to set-up digests with Time-Saver™ qualified enzymes for 5-15 minutes? Can I digest longer?
- Why is my Restriction Enzyme not cutting DNA?
- Why do I see a DNA smear on an agarose gel after a restriction digest?
- Why do I see additional DNA bands on my gel after a restriction digest?
- How many nucleotides do I have to add adjacent to the RE recognition site in order to get efficient cutting?
- Which NEB restriction enzymes are supplied with Gel Loading Dye, Purple (6X)?
- Is this enzyme sensitive to dam, dcm or mammalian CpG methylation?
- 问题解决指南
- Restriction Enzyme Troubleshooting Guide
引用 & 技术文献
- 引用文献
产品引用文献查找工具
 Powered by Bioz See more details on Bioz
Powered by Bioz See more details on Bioz
质控、安全 & 法规
- 质控分析
每一新批次的 NEB 产品都要经过质控,以满足指定的质量标准,对特定产品的质量标准和每一批次的检测数据可以通过产品说明表格、COA、产品信息卡或者产品手册进行查阅或下载。关于 NEB 产品质控的详细信息,可从此处查阅。
- 产品说明与变更通知
产品说明与变更通知
产品说明表包含产品的储存温度、有效期和规格,这些文件的命名规则如下:[货号]_[规格]_[版本]
- R0505M_v1
- R0505S_L_v1
- CoA
CoA 文件包含单个批次产品的储存温度、有效期和质控,这些文件的命名规则如下: [货号]_[规格]_[版本]_[Lot#]
- R0505S_L_v1_0511401
- R0505M_v1_0471303
- R0505M_v1_0491307
- R0505S_L_v1_0471303
- R0505S_L_v1_0491308
- R0505S_L_v1_0521410
- R0505S_L_v1_0511410
- R0505M_v1_0521411
- R0505S_L_v1_0521411
- R0505S_L_v1_0541505
- R0505S_L_v1_0521407
- R0505M_v1_0511402
- R0505M_v1_0531501
- R0505M_v1_0531505
- R0505S_L_v1_0551602
- R0505S_L_v1_0551605
- R0505M_v1_0551602
- R0505M_v1_0551608
- R0505S_L_v1_0571611
- R0505M_v1_0571709
- R0505S_L_v1_0571705
- R0505S_L_v1_0571712
- R0505M_v1_0571803
- R0505L_v1_10008867
- R0505S_v1_10010521
- R0505M_v1_10021733
- R0505L_v1_10035405
- R0505M_v1_10035407
- R0505S_v1_10035409
- R0505S_v1_10043635
- R0505L_v1_10043637
- R0505M_v1_10050279
- R0505L_v1_10065277
- R0505S_v1_10062746
- R0505M_v1_10069598
- R0505M_v1_10077743
- R0505L_v1_10084611
- R0505S_v1_10084612
- R0505L_v1_10091754
- R0505M_v1_10091766
- R0505M_v1_10092489
- R0505L_v1_10098734
- SDS
以下 SDS 文件可以帮助您安全地使用该产品
-
EagI
-
EagI |NEB酶试剂 New England Biolabs
上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
EagI
This product has been discontinued and is replaced with the High-Fidelity version, EagI-HF (NEB #R3505)
- Catalog # R0505 was discontinued on March 17, 2021
- Time-Saver™ qualified for digestion in 5-15 minutes
- High Fidelity (HF®) version available (NEB #3505) supplied with CutSmart® Buffer
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
- Restriction Enzyme Cut Site: C/GGCCG
精选视频
产品信息
EagI has a High Fidelity version, EagI-HF® (NEB #R3505)
High Fidelity (HF) Restriction Enzymes have 100% activity in CutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
产品来源
An E. coli strain that carries the EagI gene from Enterobacter agglomerans (R. Morgan).
- 产品类别:
- Discontinued Products
- 特性和用法
单位定义
One unit is defined as the amount of enzyme required to digest 1 µg of pXba DNA in 1 hour at 37°C in a total reaction volume of 50 µl.
反应条件
1X NEBuffer™ 3.1
Incubate at 37°C1X NEBuffer™ 3.1
100 mM NaCl
50 mM Tris-HCl
10 mM MgCl2
100 µg/ml BSA
(pH 7.9 @ 25°C)在不同缓冲液中的活性
NEBuffer™ 1.1: 10%
NEBuffer™ 2.1: 25%
NEBuffer™ 3.1: 100%稀释兼容性
- 稀释液 B
贮存溶液
10 mM Tris-HCl
500 mM NaCl
1 mM DTT
0.1 mM EDTA
200 µg/ml BSA
50% Glycerol
pH 8 @ 25°C热失活
65°C for 20 min
甲基化敏感性
dam 甲基化: Not Sensitive
dcm 甲基化: Not Sensitive
CpG甲基化: Blocked同裂酶
XmaIII+
- 相关产品
相关产品
- EagI-HF®
- Monarch® Plasmid Miniprep Kit
- Monarch® DNA Gel Extraction Kit
- Monarch® PCR & DNA Cleanup Kit (5 μg)
单独销售的组分
- Gel Loading Dye, Purple (6X)
- NEBuffer 3.1
- 注意事项
- EagI is an isoschizomer of XmaIII.
- For full EagI activity, the pH of the reaction mix must be between (7.9 and 9.0 @ 25°C).
- This enzyme has shown to have lower activity on some supercoiled plasmids, with more than 1 unit required to digest 1 μg plasmid DNA. For complete digestion of 1 μg of plasmid DNA please follow our recommended digestion protocol.
- Blocked by CpG methylation.
操作说明、说明书 & 用法
- 操作说明
- Optimizing Restriction Endonuclease Reactions
- Restriction Digest Protocol
- Double Digest Protocol with Standard Restriction Enzymes
- 使用指南
- Activity at 37°C for Restriction Enzymes with Alternate Incubation Temperatures
- Activity of Restriction Enzymes in PCR Buffers
- Cleavage Close to the End of DNA Fragments
- Dam and Dcm Methylases of E. coli
- Digestion of Agarose-Embedded DNA: Info for Specific Enzymes
- Double Digests
- Effects of CpG Methylation on Restriction Enzyme Cleavage
- Heat Inactivation
- NEBuffer Activity/Performance Chart with Restriction Enzymes
- Optimizing Restriction Endonuclease Reactions
- Restriction Endonucleases – Survival in a Reaction
- Restriction Enzyme Diluent Buffer Compatibility
- Restriction Enzyme Tips
- Single Letter Codes
- Star Activity
- Traditional Cloning Quick Guide
工具 & 资源
- 选择指南
- Alphabetized List of Recognition Sequences
- Cleavage of Supercoiled DNA
- Compatible Cohesive Ends and Generation of New Restriction Sites
- Dam-Dcm and CpG Methylation
- Frequencies of Restriction Sites
- Isoelectric Points (pI) for Restriction Enzymes
- Isoschizomers
- NEB Diluent and Buffer Table
- Recleavable Filled-in 5′ Overhangs
- Time-Saver™ Qualified Enzymes
- Why Choose Recombinant Enzymes?
- Web 工具
- Competitor Cross-Reference Tool
- DNA Sequences and Maps Tool
- Double Digest Finder
- Enzyme Finder
- NEBcutter™ v3.0
- NEBioCalculator®
- REBASE®
FAQs & 问题解决指南
- FAQs
- Does EagI produce commonly used compatible ends?
- Are more units of EagI required to cut supercoiled DNA than lambda DNA?
- Do I have to set-up digests with Time-Saver™ qualified enzymes for 5-15 minutes? Can I digest longer?
- Why is my Restriction Enzyme not cutting DNA?
- Why do I see a DNA smear on an agarose gel after a restriction digest?
- Why do I see additional DNA bands on my gel after a restriction digest?
- How many nucleotides do I have to add adjacent to the RE recognition site in order to get efficient cutting?
- Which NEB restriction enzymes are supplied with Gel Loading Dye, Purple (6X)?
- Is this enzyme sensitive to dam, dcm or mammalian CpG methylation?
- 问题解决指南
- Restriction Enzyme Troubleshooting Guide
引用 & 技术文献
- 引用文献
产品引用文献查找工具
 Powered by Bioz See more details on Bioz
Powered by Bioz See more details on Bioz
质控、安全 & 法规
- 质控分析
每一新批次的 NEB 产品都要经过质控,以满足指定的质量标准,对特定产品的质量标准和每一批次的检测数据可以通过产品说明表格、COA、产品信息卡或者产品手册进行查阅或下载。关于 NEB 产品质控的详细信息,可从此处查阅。
- 产品说明与变更通知
产品说明与变更通知
产品说明表包含产品的储存温度、有效期和规格,这些文件的命名规则如下:[货号]_[规格]_[版本]
- R0505M_v1
- R0505S_L_v1
- CoA
CoA 文件包含单个批次产品的储存温度、有效期和质控,这些文件的命名规则如下: [货号]_[规格]_[版本]_[Lot#]
- R0505S_L_v1_0511401
- R0505M_v1_0471303
- R0505M_v1_0491307
- R0505S_L_v1_0471303
- R0505S_L_v1_0491308
- R0505S_L_v1_0521410
- R0505S_L_v1_0511410
- R0505M_v1_0521411
- R0505S_L_v1_0521411
- R0505S_L_v1_0541505
- R0505S_L_v1_0521407
- R0505M_v1_0511402
- R0505M_v1_0531501
- R0505M_v1_0531505
- R0505S_L_v1_0551602
- R0505S_L_v1_0551605
- R0505M_v1_0551602
- R0505M_v1_0551608
- R0505S_L_v1_0571611
- R0505M_v1_0571709
- R0505S_L_v1_0571705
- R0505S_L_v1_0571712
- R0505M_v1_0571803
- R0505L_v1_10008867
- R0505S_v1_10010521
- R0505M_v1_10021733
- R0505L_v1_10035405
- R0505M_v1_10035407
- R0505S_v1_10035409
- R0505S_v1_10043635
- R0505L_v1_10043637
- R0505M_v1_10050279
- R0505L_v1_10065277
- R0505S_v1_10062746
- R0505M_v1_10069598
- R0505M_v1_10077743
- R0505L_v1_10084611
- R0505S_v1_10084612
- R0505L_v1_10091754
- R0505M_v1_10091766
- R0505M_v1_10092489
- R0505L_v1_10098734
- SDS
以下 SDS 文件可以帮助您安全地使用该产品
-
EagI
-
NEB酶试剂 New England Biolabsuffer 1.1 | NEB酶试剂 New England Biolabs
上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
NEBuffer™ 1.1
This product will be discontinued on 12/15/2021 and replaced with NEBuffer™ Set (r1.1, r2.1, r3.1 and rCutSmart™.
- Catalog # B7201 was discontinued on December 15, 2021
Most of our enzymes are supplied with one of four standard NEBuffers. Visit NEBCutSmart.com for details.

产品信息
New England Biolabs provides a colorcoded 10X NEBuffer with each restriction endonuclease to ensure optimal (100%) activity. Most of our enzymes are supplied with one of four standard NEBuffers. Occasionally, an enzyme has specific buffer requirements not met by one of the four standard NEBuffers, in which case the enzyme is supplied with its own unique NEBuffer.
- 产品类别:
- Discontinued Products
- 特性和用法
1X 缓冲液组分
10 mM Bis-Tris-Propane-HCl
10 mM MgCl2
100 µg/ml BSA
pH [email protected]°C - 相关产品
相关产品
- Monarch® Plasmid Miniprep Kit
- Monarch® DNA Gel Extraction Kit
- Monarch® PCR & DNA Cleanup Kit (5 μg)
- 注意事项
- The pH of 10X NEBuffer 1.1 is between pH 6.9 and 7.1.
工具 & 资源
- 选择指南
- NEB Diluent and Buffer Table
FAQs & 问题解决指南
- FAQs
- What is supplied with the NEBuffer 1.1 pack?
- How should the NEBuffer be used?
- The buffer arrived thawed. Are the buffer and the enzyme still active?
- You have replaced NEBuffer 1 with NEbuffer 1.1, NEBuffer 2 with NEBuffer 2.1, and NEBuffer 3 with 3.1. Where is NEBuffer 4.1?
- Why did you add BSA into all the restriction enzyme reaction buffers?
- Why did you remove DTT from your restriction enzyme buffers?
- I don’t want to use BSA in my buffer. What are my options?
- Why is my Restriction Enzyme not cutting DNA?
- Why do I see additional DNA bands on my gel after a restriction digest?
- Why do I see a DNA smear on an agarose gel after a restriction digest?
- How many nucleotides do I have to add adjacent to the RE recognition site in order to get efficient cutting?
- Are the alkaline phosphatases active in NEBuffers?
引用 & 技术文献
- 引用文献
产品引用文献查找工具
 Powered by Bioz See more details on Bioz
Powered by Bioz See more details on Bioz
质控、安全 & 法规
- 质控分析
每一新批次的 NEB 产品都要经过质控,以满足指定的质量标准,对特定产品的质量标准和每一批次的检测数据可以通过产品说明表格、COA、产品信息卡或者产品手册进行查阅或下载。关于 NEB 产品质控的详细信息,可从此处查阅。
- 产品说明与变更通知
产品说明与变更通知
产品说明表包含产品的储存温度、有效期和规格,这些文件的命名规则如下:[货号]_[规格]_[版本]
- B7201S_v1
- CoA
CoA 文件包含单个批次产品的储存温度、有效期和质控,这些文件的命名规则如下: [货号]_[规格]_[版本]_[Lot#]
- B7201S_v1_0131803
- B7201S_v1_10010011
- B7201S_v1_10033676
- B7201S_v1_10046907
- B7201S_v1_10048363
- B7201S_v1_10055575
- B7201S_v1_10057224
- B7201S_v1_10090430
- B7201S_v1_10093183
- B7201S_v1_10102472
- SDS
以下 SDS 文件可以帮助您安全地使用该产品
-
NEBuffer™ 1.1
-